Biomarkers: predicting response to treatment
Better PFS with CAPTEM in MEN1-mut/DAXX-wt pNET patients
pNETs frequently contain mutations in MEN1, ATRX, DAXX, and the PI3K/AKT/mTOR pathway [1]. However, more data are needed to determine whether this information can predict response to standard treatments, such as CAPTEM.
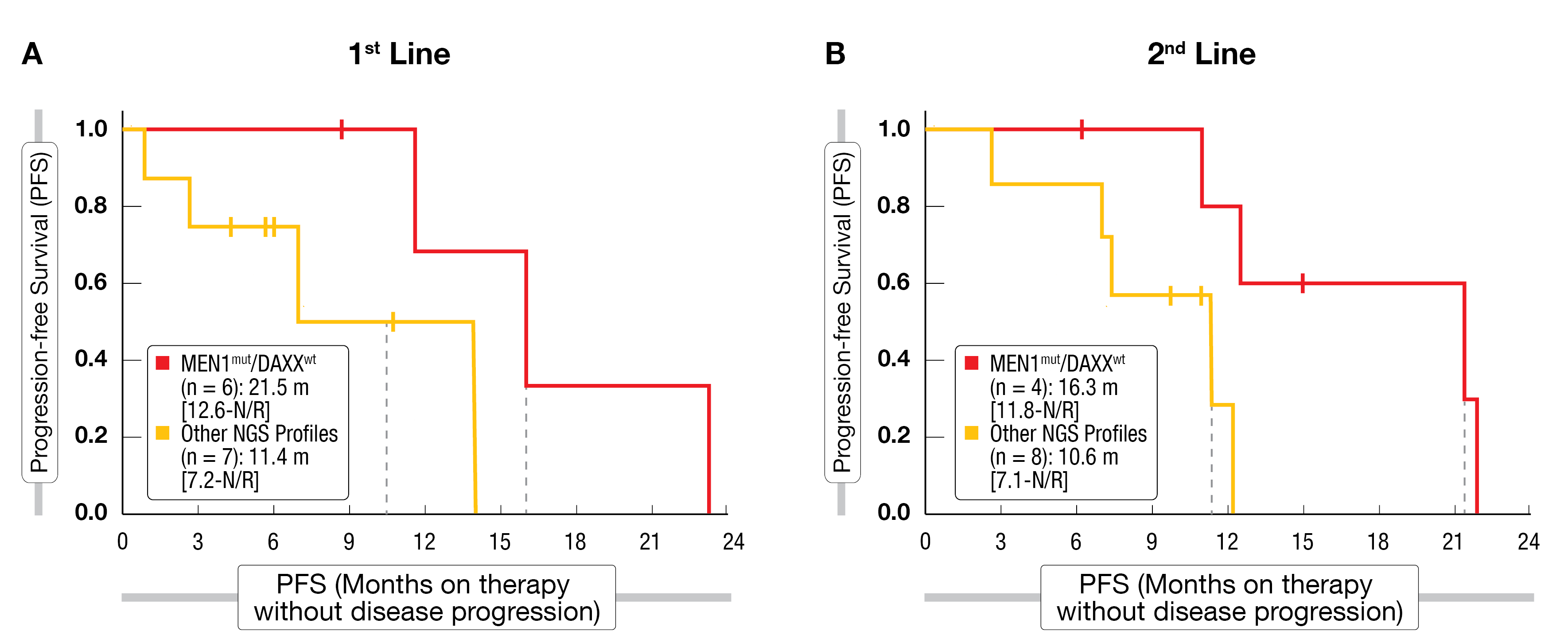
At NANETS 2022, Hendifar et al. presented retrospective data on 25 patients with well-differentiated grade 1 and 2 pNETs who had received CAPTEM as first- or second-line treatment and whose tumors had been molecularly characterized through next-generation sequencing (NGS). As reported by the authors, MEN1 mutations were positively associated with CAPTEM response. However, this effect was less pronounced in the subset of patients with co-occurring DAXX mutations, which are commonly found together with MEN1 alterations. PFS with CAPTEM was significantly longer, regardless of line of therapy, in MEN1-mut/DAXX-wt pNET patients compared to other genomic profiles (Figure 1) [2]. The correlation of this novel genomic signature with response to CAPTEM in pNETs should be validated in a prospective and larger cohort.
Figure 1: Progression-free survival outcomes with first (A) or second (B) line CAPTEM in pNET subgroups defined by their MEN1/DAXX mutational status
PRRT-predictive quotient predicts response to PRRT
Another treatment strategy commonly used in the management of NETs is PRRT [3]. However, there is also a lack of reliable molecular biomarkers predicting its clinical efficacy. Data regarding a PRRT-predictive-quotient (PPQ) was presented at NANETS 2022 and may be a promising non-invasive tool to improve the management of NET patients undergoing PRRT.
PPQ is a blood-based genomic assay that integrates circulating levels of NET-specific gene transcripts with tumor grade (as determined by Ki67 staining) to provide information on tumor radiosensitivity and PRRT responsiveness (PPQ+ = predicted PRRT responder). In a previous study on three independent cohorts of 177Lu-PRRT-treated lung and GEP-NET patients, PPQ was shown to predict response to PRRT in a specific manner and with an accuracy of >95 % (compared to an accuracy of only 50 % in SSA-treated cohorts) [4, 5]. The efficacy of PPQ as a predictive marker of PRRT response is currently being validated in a prospective cohort of metastatic NET patients in the US, where the potential of PPQ to predict the toxicity of PRRT will also be evaluated [6].
Immune-related effects as biomarkers of SSA response
Treatment with SSAs such as lanreotide is considered standard of care in advanced NETs. SSAs exert an inhibitory effect on tumor cells through binding to the overexpressed SSTR2 [7]. However, the effect of SSAs on immune cells, which have also been shown to differentially express SSTR1-5, is not well understood.
To identify potential biomarkers that would predict response to SSA, Maguire et al. investigated the effects of lanreotide on different subsets of T-cells sorted from a cohort of SSA-treated NET patients (9 responders and 8 non-responders). According to the results from gene and protein expression analysis presented at NANETS 2022, a basal (pre-treatment) upregulation of T-cell receptor and interferon signaling in CD4+ and CD8+ cells, respectively, indicated a greater immunological competence of responders compared to non-responders. Three months after SSA treatment, downregulation of cytokine and chemokine signaling as well as upregulation of ubiquitination and proteasome degradation-associated genes was observed in responders [8]. This study underscored the relevance of immune effects associated with SSA therapy. Based on these data, the clinical utility of the differentially expressed genes in responders vs. non-responders as predictive markers of SSA response may be determined.
New strategies for molecular profiling of NETs
Optical genome mapping (OGM) was presented at NANETS 2022 as a novel method for the identification of genomic structural variants that may drive the progression of metastatic NETs [9]. NETs have generally been thought to have a low mutation burden [1, 10]. Considering that genetic profiling is usually performed with short-read sequencing technologies, which are able to detect single nucleotide variants but might miss large structural variants [11], the addition of OGM would provide complementary information and contribute to a better molecular characterization of NETs.
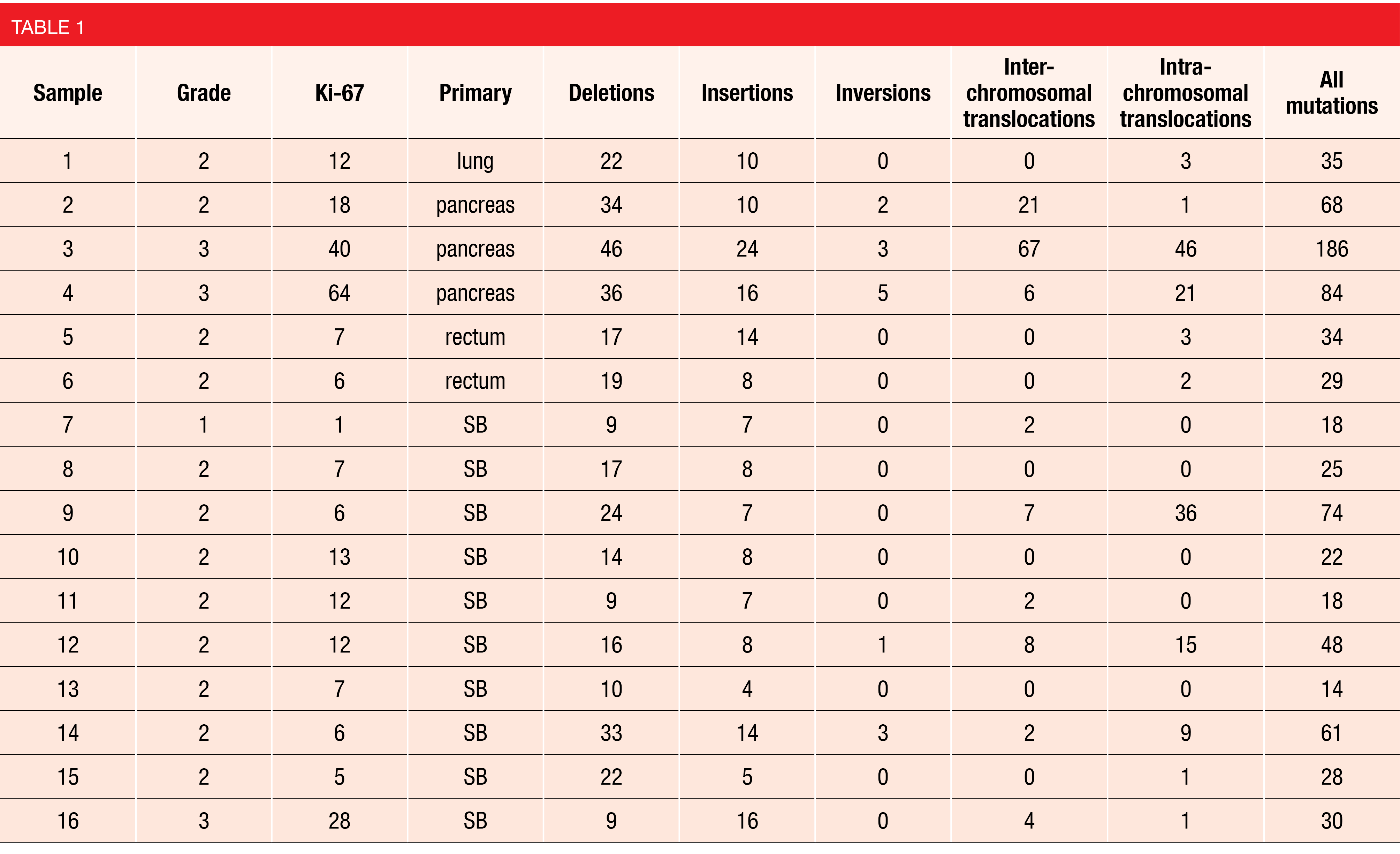
A proof-of-concept study evaluated OGM using biopsy samples from 16 metastatic NET patients of different grade and primary site. The authors reported a mean of 48 ± 43 and a median of 32 structural variants identified in each sample. On average, deletions accounted for 44 % of variants, insertions for 21 %, inversions for 2 %, inter-chromosomal translocations for 15 %, and intra-chromosomal translocations for 18 % (Table 1). OGM not only identified structural variants in all samples, but also found trends towards differences between tumors of different grade and primary site of origin [9]. New variants identified by OGM could be further correlated with clinical data and outcomes to identify potential biomarkers that may help improve the clinical management of NETs.
An alternative strategy for the molecular profiling of pNENs that may help provide better tailored treatments for these patients was presented by Lou et al. Based on an analysis of 318 NEN cases from which histological grade annotation as well as NGS and WTS data were available, a threshold of MKI67 expression able to differentiate low grade (LG) from high grade (HG)NENs was defined. This MKI67 threshold was then validated in a larger cohort of NEN patients (n = 1768). The differences between the mutational landscapes of HG- vs. LG-NENs observed in pathology-based cohorts were recapitulated in the HG and LG cohorts inferred from MKI67 expression, including TP53, KRAS and RB1. These molecular alterations were more frequent in HG-NENs than in LG (∆ prevalence = 44.31 %, 24.09 % and 25.61 %, respectively; q < 0.05), while LG-NENs were found to have higher expression of SSTR1-3 (1.12-fold, 1.77-fold and 1.06-fold, respectively; q < 0.05). In contrast, SSTR4 was significantly higher in HG-NENs (3.87-fold, q < 0.05). Subsequently, the prevalence of each mutation was assessed according to the expression levels of each SSTR subtype. In HG-NENs, MEN1, ATRX and TSC2 were increased among SSTR1-2-high cases, while KRAS and RB1 were more frequent in SSTR1-2-low tumors [12]. The authors concluded that transcriptomics could be leveraged to predict pNEN grade and to identify the molecular profiles associated with the expression of each SSTR subtype. Thus, routinely assessing SSTR subtype expression and incorporating molecular profiling might help to improve clinical decision-making, e.g., the identification of patients eligible for PRRT among other therapies.
REFERENCES
- Di Domenico A et al., Genetic and epigenetic drivers of neuroendocrine tumours (NET). Endocr Relat Cancer 2017; 24(9): R315-R334
- Lee PC et al., Correlation of MEN1 and DAXX mutational status with response to capecitabine and temozolomide (CAPTEM) in pancreatic neuroendocrine tumors. NANETS 2022, abstract C-9
- Hope TA et al., NANETS/SNMMI Procedure Standard for Somatostatin Receptor-Based Peptide Receptor Radionuclide Therapy with 177Lu-DOTATATE. J Nucl Med 2019; 60(7): 937-943.
- Bodei L et al., PRRT neuroendocrine tumor response monitored using circulating transcript analysis: the NETest. Eur J Nucl Med Mol Imaging 2020; 47(4): 895-906
- Bodei L et al., PRRT genomic signature in blood for prediction of 177Lu-octreotate efficacy. Eur J Nucl Med Mol Imaging 2018; 45(7): 1155-1169
- Bodei L et al., Interim analysis of a prospective validation of two blood-based genomic assessments (PPQ and NETest) to determine clinical efficacy of 177Lu-DOTATATE in neuroendocrine tumors. J Nucl Med 2022; jnumed.122.264363
- Caplin ME et al., Lanreotide in metastatic enteropancreatic neuroendocrine tumors. N Engl J Med 2014; 371(3): 224-233
- Alaklabi S et al., Immune cell molecular pharmacodynamics of lanreotide in relation to treatment response. NANETS 2022, abstract O-4
- DePietro DM et al., Optical genome mapping: a novel approach to identifying structural variants in metastatic neuroendocrine tumors. NANETS 2022, abstract B-13
- Banck MS et al., The genomic landscape of small intestine neuroendocrine tumors. J Clin Invest 2013; 123(6): 2502-8
- Huddleston J et al., Discovery and genotyping of structural variation from long-read haploid genome sequence data. Genome Res 2017; 27(5): 677-685
- Lou E et al., Leveraging transcriptomics to grade pancreatic neuroendocrine neoplasms (NENs) and assess molecular alterations associated with somatostatin receptor (SSTR) subtype expression. NANETS 2022, abstract O-9
© 2023 Springer-Verlag GmbH, Impressum
More posts
Preface – NANETS 2022
Preface – NANETS 2022 © private - Mauro Cives, MD, Department of Interdisciplinary Me