Focus Molecular Diagnostics
Molecular diagnostics: which markers, which methods?
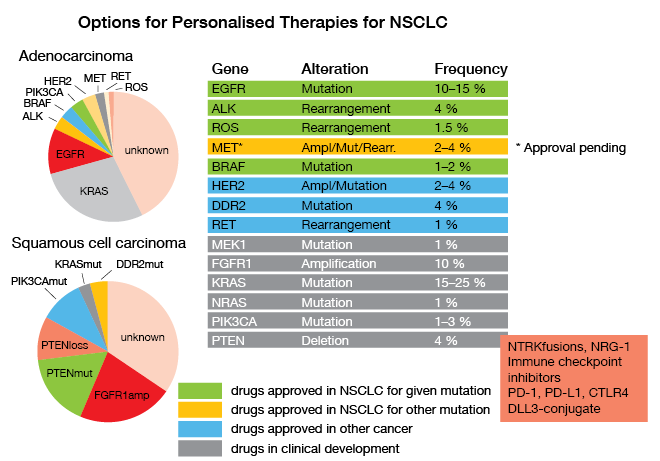
All existing guidelines call for timely determination of molecular parameters relevant for approved targeted and immune treatments of unresectable NSCLC, as Reinhard Büttner, MD, Department of Pathology, University Hospital of Cologne, emphasized. EGFR mutations, ALK and ROS rearrangements, BRAF mutations, PD-L1 expression and – hopefully soon – amplifications, mutations or rearrangements of MET have been connected with effective new therapeutics, and have therefore to be identified as soon as possible in the course of disease (Fig. 6). This is not only true for first-line therapy, but also in the resistant situation where molecular pathology is also involved in decision making; e. g., when detection of a T790M-mutation in the EGFR gene opens the way to treatment with the third-generation inhibitor osimertinib, or when resistance mutations in the ALK-fusion gene dictate the choice of ALK inhibitors. So, there are roughly 27 genes and a series of immunohistochemically determined parameters that have to be analyzed for the first-line and second-line situations, to ascertain that the patients are treated with the extremely successful therapies that target the respective alterations.
Another strategy, which in recent years has been employed very successfully for treatment of many tumors including NSCLC, is the blockade of immune checkpoint molecules like PD-1. The immune checkpoint mechanisms that have developed in the mammalian immune system have the task of protecting tissues from autoimmune attacks by T-cells – in utero as well as in a variety of immune-privileged spaces in the adult organism. Checkpoint molecules like PD-1 on T-cells recognize specific ligands like PD-L1 on tissue cells, and as a consequence of this interaction the cytotoxic activity of the immune cells is being blocked. Cancer cells utilize this mechanism by expressing proteins like PD-L1 themselves and thereby preventing their own lysis by T-cells. In the KEYNOTE-024 trial first-line therapy with the PD-1 antibody pembrolizumab could prolong PFS as well as OS of patients with NSCLC expressing PD-L1 on at least 50 % of their cells as determined by IHCs [1].
Determination of PD-L1 expression is therefore of paramount importance when considering checkpoint inhibitor therapies for patients with NSCLC. At the moment, PD-L1 expression is the only approved biomarker for immunotherapy, and for first-line treatment with pembrolizumab, it is mandatory to score the proportion of tumor cells that express PD-L1; in addition, it is optional to determine the proportion of PD-L1–positive immune cells. Andreas Scheel, MD, Department of Pathology, University Hospital of Cologne, led the pivotal German harmonization trial for standardizing PD-L1 IHC [2–4]. This IHC technique is a fast and relatively cheap method, and it requires little tissue. It can be standardized to a high degree, although appropriate validation and quality control are essential. Furthermore, each of the five PD-1/PD-L1 inhibitors has been validated clinically with different IHC assays, which are not interchangeable. Biomarker testing will certainly develop further, Scheel stated, because more clinical trials with different PD-L1 IHC tests and cut-offs are ongoing, and more methods are being evaluated, including RNA expression analysis and comprehensive DNA sequencing, to define the so-called ‘tumor mutational burden’.
Traditionally molecular genetics testing has been performed using multiplex PCR, with specific panels for each type of tumor, thereby restricting the region of DNA investigated to a few thousand base pairs. This is currently not enough for assumptions to be made concerning the total mutational burden (e.g., microsatellite instability, BRCA mutation, UV or smoking signature). So, more recently in Cologne, larger hybrid capture panels were developed that allow determination of mutations, fusions and amplifications, and also patterns of mutational load that include copy-number variations [5].
Indeed, this type of assay might explain the contradictory results from some checkpoint inhibitor trials, where some of the patients with high PD-L1 expression did not respond. In the CheckMate-026 trial, for example, patients with PD-L1 expression > 50 % but with low or medium tumor mutational burden had shorter PFS with nivolumab than with chemotherapy, while for those with PD-L1 > 50 % and high tumor mutational burden, the median PFS was not reached after 18 months. Measurement of tumor mutational burden therefore is a very promising biomarker candidate, provided there is an assay that can be used readily in clinical practice. Usually, the tumor mutational burden has been determined using whole exome sequencing, which, however, requires at least 200 ng DNA from formalin-fixed, paraffin-embedded tissues. For example, in the CheckMate-026 trial, sufficient samples could not be obtained from 42 % of the patients. Therefore, there are efforts underway, as Sabine Merkelbach-Bruse, MD, Department of Pathology, University Hospital of Cologne, stated, to develop panel sequencing techniques for tumor mutational burden analysis.
In addition, the nature of the immune infiltrate of a tumor might be relevant for patient prognosis, such as whether a tumor consists predominantly of T-cells or of more immunosuppressive myeloid cells. The aim, as Büttner put it, is the creation of an “integrated immune score”, as some cancer immunologists called it a couple of years ago [6].
Figure 6: Options for Personalised Therapies for NSCLC
Implementation: Network Genomics Medicine
When talking about the Network of Genomics Medicine (NGM) that was founded at the University of Cologne in 2010, Anna Kron, University Hospital of Cologne, mentioned its association with the Lung Cancer Group Cologne (LCGC) to achieve a number of challenging goals:
- establish a comprehensive clinical trials program (with a focus on early proof-of-concept trials);
- participate in (or better, lead) practice-changing pharmaceutical trials;
- translate discoveries from the academic setting into clinical practice (‘from bench to bedside’);
- initiate investigator-initiated trials within LCGC, and expand them to multicenter trials;
- start an immunotherapy trial program, and try to integrate this with genomics (BIOLUMA).
The first evaluation of the NGM in 2013 already showed clear superiority over standard chemotherapy of personalized treatments with EGFR and ALK inhibitors after molecular testing, with these data in line with those from the respective controlled trials [7]. At present, the NGM provides genotyping of lung cancers for about 10 % of all patients in Germany, with the ultimate goal being to increase this to 100 %. Reimbursement of next-generation sequencing is now possible for around one third of all lung cancer patients in Germany, with the final goal being uniform cost coverage for all in-patients and out-patients. Having repeatedly shown survival benefits through molecular testing and participation in clinical trials, the NGM is striving to achieve nationwide documentation and evaluation of treatments and outcomes in patients with lung cancer. Nationwide molecular screening is another goal, with one purpose (among others) being the organization of clinical trials for rare subgroups of patients. Finally, Merkelbach-Bruse mentioned the harmonization of quality standards for diagnostics and treatment as the overarching aim of all of these efforts, one prominent example being the German harmonization study for PD-L1 testing.
To achieve all of this at a national level, the National NGM was founded in September 2017, and funding from German Cancer Aid started on April 1, 2018. In this context, lung cancer serves as a model for other solid tumors.
The main reason for implementation of molecular diagnostics as a tool in daily practice lies in the need to better differentiate groups of patients with a diagnosis of lung cancer. The organizational investments are huge, but they are justified by the ever-growing complexity of the individual therapies that are mainly determined by patient characteristics, molecular markers, and new substances, and the success that can be achieved by this approach that has been shown in clinical trials.
REFERENCES
- Reck M et al. Pembrolizumab versus chemotherapy for PD-L1-positive non–small-cell lung cancer. N Engl J Med 2016; 375: 1823-33.
- Scheel AH et al. Harmonized PD-L1 immunohistochemistry for pulmonary squamous-cell and adenocarcinomas. Mod Pathol 2016; 29: 1165-72.
- Scheel AH et al. Interlaboratory concordance of PD-L1 immunohistochemistry for non–small-cell lung cancer. Histopathololy 2017; 72: 449-59.
- Büttner R et al. Programmed death-ligand 1 immunohistochemistry testing: a review of analytical assays and clinical implementation in non–small-cell lung cancer. J Clin Oncol 2017; 35: 3867-76.
- Heydt C et al. ALK evaluation in the world of multiplex testing: Network Genomic Medicine (NGM): the Cologne model for implementing personalised oncology. Ann Oncol 2016; 27 (Suppl 3): iii25-iii34.
- Blank CU et al. Cancer Immunology. The ‘cancer immunogram’. Science 2016; 352: 658-60.
- The Clinical Lung Cancer Genome Project and Network Genomic Medicine. A genomics-based classification of human lung tumors. Sci Transl Med 2013; 5: 209ra153.