Precision medicine in the field of lung cancer in Japan
In 2013, Japanese researchers launched a nationwide, prospective, observational study for patients with lung cancer and genomic alterations, called Lung Cancer Genomic Screening Project for Individualized Medicine in Japan (LC-SCRUM-Japan). The number of participating institutions has risen greatly over time, and today 241 institutions from almost all regions across the country are part of the project.
The tumors of patients with locally advanced or metastatic NSCLC who participate in LC-SCRUM are tested centrally for genomic alterations. Testing is based on fresh frozen samples plus formalin-fixed, paraffin-embedded (FFPE) tissue, or pleural effusion. Until 2015, real-time PCR (RT-PCR) and fluorescence in situ hybridization (FISH) have been the technique of choice for non-squamous NSCLC, but it has been replaced by multiplex genome analysis using next-generation sequencing (NGS). If testing reveals an actionable aberration, the patient enters a trial. These studies can be either investigator-initiated or company-initiated. The program has expanded greatly; initially, it only covered patients with non-squamous, EGFR-mutation–negative NSCLC, but today there are also distinct projects for squamous NSCLC, small-cell tumors, and for biomarker assessment in the immuno-oncological setting (LC-SCRUM-Japan IBIS). Also, patients with unknown EGFR mutation status started to be accepted into the screening program.
Screening results
As of 30th September 2017, 4,820 patients have been tested in the LC-SCRUM-Japan project. Fresh frozen samples plus FFPE constituted the majority of samples. More than 90 % of these were adequate for both RT-PCR and NGS. Out of 3,394 samples that were tested using RT-PCR or NGS until the end of August 2017, 4 %, 3 % and 2 % tested positive for ROS1, RET, and ALK fusion, respectively. Ninety-one percent of the other samples were negative for these three fusion genes as well as for EGFR mutation.
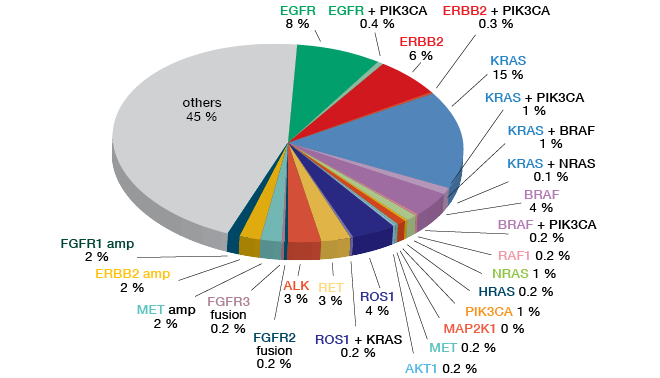
Testing of non-squamous NSCLC by means of either the OncomineTM Cancer Research Panel (OCP) (Figure 4) or the OncomineTM Comprehensive Assay (OCA) showed a range of established aberrations, such as EGFR mutation, HER2 mutation, BRAF mutation, ALK fusion, and FGFR1 amplification. Three percent of patients included in LC-SCRUM-Japan were positive for MET exon 14 skipping mutation.
Screening results for squamous cell lung cancer using OCP revealed that FGFR and PIK3CA aberrations were prevalent here, as well as KRAS mutations. For tumors with small-cell histology, the proportion of identifiable aberrations was quite low. Alterations included PIK3CA mutation, PTEN mutation, MYC, MYCL1 and MYCN amplification, EGFR mutation, FGFR1 gain, KRAS mutation, and TSC2 mutation. Eight percent of the patients showed PI3K/AKT/mTOR pathway mutations.
Figure 4: Screening results obtained in 1,688 non-squamous NSCLC samples collected in LC-SCRUM-Japan
LURET and other trials
Patients who tested positive for RET rearrangement according to both RT-PCR and FISH were included in the phase II LURET study that assessed the efficacy of the oral RET inhibitor vandetanib at a dose of 300 mg/d after at least one prior chemotherapy. Among 1,433 screened patients, 34 patients tested RET-positive, and 19 entered the trial including 2 ineligible patients [1]. Objective response rate (ORR) constituted the primary endpoint. The study was positive, with an ORR of 53 % (90 % CI: 31-74 %) (Figure 5).
To date, a total of 653 NSCLC patients with actionable gene alterations have been screened for clinical trials. Among these, 174 had HER2 amplification/ mutation, 135 had ROS1 fusion, and 96 were shown to harbor RET fusion. ALK fusion was present in 78 patients, MET amplification/ exon 14 skipping mutation in 76, PIK3CA mutation in 52, and NTRK3 fusion in 1 patient. Overall, 128 patients have entered clinical trials. Both well-known agents (e.g., crizotinib, alectinib) and experimental drugs are evaluated. For some alterations, multiple treatment approaches exist; patients with RET fusion, for instance, have entered studies testing vandetanib, lenvatinib, and alectinib. Twenty-five clinical trials targeting genomic alterations that have been identified in LC-SCRUM-Japan are ongoing or have been conducted to date.
Figure 5: LURET trial: responses to vantedanib in patients with RET-rearranged NSCLC.
Modified from Yoh K et al. [1]
SCRUM-Japan
The next step after the inception of LC-SCRUM-Japan was the implementation of SCRUM-Japan, a nationwide cancer genome screening project which covers entities beyond lung cancer. It is based on collaborations between academic institutions, the industry and the government; funding is provided by 16 pharmaceutical companies as well as through grants from the Japanese Agency for Medical Research and Development (AMED) and the National Cancer Center. Public databases can be used to conduct research.
Delivering appropriate therapeutic agents to each cancer patient is at the heart of SCRUM-Japan. This includes the nationwide cancer genome screening for orphan-fractionated cancers, the clinical development of corresponding therapeutic products based on cancer genome alterations, and a strong support for the clinical development of companion diagnostics such as NGS multiplex genomic diagnostics.
REFERENCE
- Yoh K et al., Vandetanib in patients with previously treated RET-rearranged advanced non-small-cell lung cancer (LURET): an open-label, multicentre phase 2 trial. Lancet Respir Med 2017; 5(1): 42-50


![Figure 5: LURET trial: responses to vantedanib in patients with RET-rearranged NSCLC. Modified from Yoh K et al. [1]](https://memoinoncology.com/wp-content/uploads/2020/04/Grafik-12-preceptorship-singapore-en.png)