Liquid biopsy in the context of EGFR and other mutations
Compared to tissue biopsy and re-biopsy, liquid biopsy offers several advantages, including minimal-invasiveness, the opportunity for serial measurements over time to monitor tumour response, and detection of resistance mutations in the plasma prior to radiographic detection [1]. The issue of tumour heterogeneity, which is an important factor in therapeutic failure, is also considered. Driver mutations can be identified with high sensitivity and specificity, thus improving delivery of personalised medicine. Although there remain controversial issues such as standardisation, validation of different technologies, and concordance with tissue molecular profile results, liquid biopsy has emerged as an alternative tool for the management of advanced NSCLC patients.
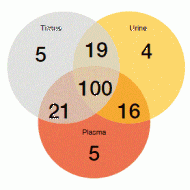
Figure: Increase in T790M detection with combined use of urine and plasma testing (170 T790M-positive cases)
High concordance rate between plasma and tissue
One of several analyses presented at the WCLC that confirm liquid biopsy as an emerging standard was that of Mack et al., who assessed the Guardant360 panel for population-scale genomics (in comparison with The Cancer Genome Atlas), clinical accuracy, and clinical utility [2]. Guardant360 testing allows for digital sequencing of critical exons in 73 genes based on circulating tumour DNA (ctDNA). The cohort comprised 8,388 patients with stage III/IV adenocarcinoma (n = 4,142) or NSCLC-NOS (n = 4,246), with 9,202 samples taken. A median time-span of 177 days had passed between initial diagnosis and ctDNA collection. Tissue information was available in a subset of patients. It should be noted that this was not a random cross-section of patients, as the analysis was enriched for patients progressing on targeted agents. They were generally being treated in the second or later lines.
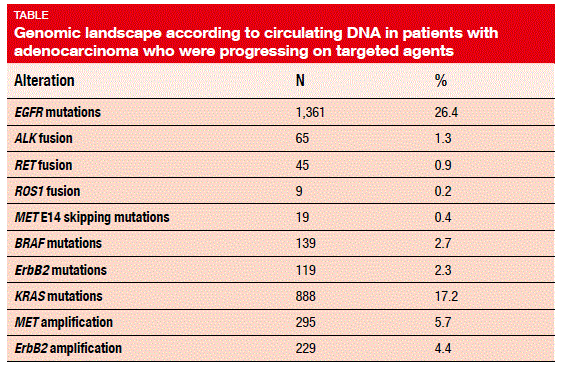
The overall detection rate of alterations was 87 %, with a median number of three alterations per sample (range, 0-93). Mutations detected in the plasma showed similar frequency and distribution as those reported in the tissue, which applied to truncal mutations present in all lineages of the tumour. The ctDNA fusion patterns mirrored tumour tissue, according to Guardant360. In patients with adenocarcinoma, EGFR mutations were found in 26.4 % of cases (Table). Exon 19 deletions constituted most of the EGFR driver mutations (52 %), followed by L858R mutations (34 %) and exon 20 insertions (4 %).
As already known, driver mutations were mutually exclusive to a statistically highly signifi cant degree. For instance, when EGFR mutation was present, KRAS mutation was not, and vice versa. Cases of overlap might be due to the emergence of secondary resistance mutations.
Increase in biomarker yield of 65 %
Clinical accuracy was determined in a subset of 543 marker-positive cases where tissue information was available. Here, positive predictive values ranged between 92 % and 100 % according to the type of mutation. All of the patients with positive plasma samples for KRAS, BRAFV600E and MET E14 skipping mutations also had these mutations in their tumour tissue. For ALK, RET and ROS1 fusions, 92 % did not show positive tissue results; these were most likely false negatives. Forty percent of ALK fusion cases and 50 % of EGFR-positive cases had one potentially actionable resistance target at progression. Overall, the plasma analysis conferred additional benefi t, as ctDNA next-generation sequencing increased the biomarker yield by 65 %. Th is corresponded to 252 additional actionable biomarkers. Oncogenic drivers were detected in 29 % of cases of under-genotyped or unevaluable tissue.
Santos et al. also used Guardant360 testing for liquid biopsy assessment in 100 consecutive patients with stage IV or recurrent adenocarcinoma [3]. Tissue molecular profi le results were obtained or recovered from each subject for purposes of comparison with their liquid biopsy counterparts. The investigators showed that agreement between the two methodologies with regard to the type of aberration was greatest for EGFR mutations (68 %). This was the case even though circulating DNA testing had been performed months or even years after tumour tissue testing. None of the liquid biopsies was performed at the time of diagnosis or tumour biopsy.
The rate of identification of abnormalities was higher with liquid biopsy than with tissue testing. Forty-six percent of patients with EGFR aberrations according to liquid biopsy had actionable mutations. Sixteen out of 35 patients with EGFR alterations showed mutations or variants identified by liquid biopsy only; in 5 of these 16 cases, actionable EGFR mutants were identified exclusively by use of liquid biopsy.
T790M mutation detection
In the TIGER-X phase I/II trial, combined EGFR mutation testing of urine and plasma was performed and analysed [4]. TIGER-X enrolled 548 patients with activating EGFR mutations who had already been treated with EGFR-directed TKIs. They received the EGFR TKI rociletinib, which is no longer in clinical development. In this trial, 540 tissue samples, 482 plasma samples and 213 urine samples were submitted for pre-treatment EGFR testing. The analysis contained 174 matched tissue, plasma and urine samples.
Non-invasive urine and plasma T790M detection was demonstrated to be highly sensitive. For both plasma and urine testing, sensitivity exceeded 80 %. Even higher rates occurred for combined testing, where the sensitivity was 96.6 %. For the 174 matched tissue, plasma and urine specimens, T790M positivity by any one specimen type was 97.7 %. Combined urine and plasma testing identifi ed more T790M-positive cases than tissue testing alone (Figure). Response rates observed with rociletinib were similar, regardless of whether T790M mutations were detected by liquid biopsy or tissue biopsy.
Moreover, the analyses showed that T790M mutations were more readily detected in the plasma of patients with extrathoracic lesions (M1b) than in those who had only intrathoracic (M1a/M0) disease. However, combined urine and plasma testing allowed for sensitive detection regardless of disease state. Sensitivity was 90.7 % and 95.8 % in patients with M1a/M0 and M1b disease, respectively. The authors concluded that the combined analysis of urine and plasma should be considered prior to tissue testing in EGFR-TKI-resistant NSCLC patients, including those with extrathoracic metastases.
REFERENCES
- Burrell RA & Swanton C, Tumour heterogeneity and the evolution of polyclonal drug resistance. Mol Oncol 2014; 8(6): 1095-1111
- Mack PC et al., Clinical utility of circulating tumor DNA (ctDNA) analysis by digital next generation sequencing of over 5,000 advanced NSCLC patients. WCLC 2016, OA06.01
- Santos ES et al., Report on liquid biopsies from advanced lung adenocarcinoma patients and correlation with their tumor biopsy profiles. WCLC 2016, OA10.07
- Wakelee HA et al., A highly sensitive nextgeneration sequencing platform for detection of NSCLC EGFR T790M mutation in urine and plasma. WCLC 2016, MA08.01